Getting Started¶
This document will show you how to install and run Scikit-ribo.
What is Scikit-ribo¶
Scikit-ribo is an open-source software for accurate genome-wide A-site prediction and translation efficiency inference from Riboseq and RNAseq data.
Source Code: https://github.com/hanfang/scikit-ribo
Introduction¶
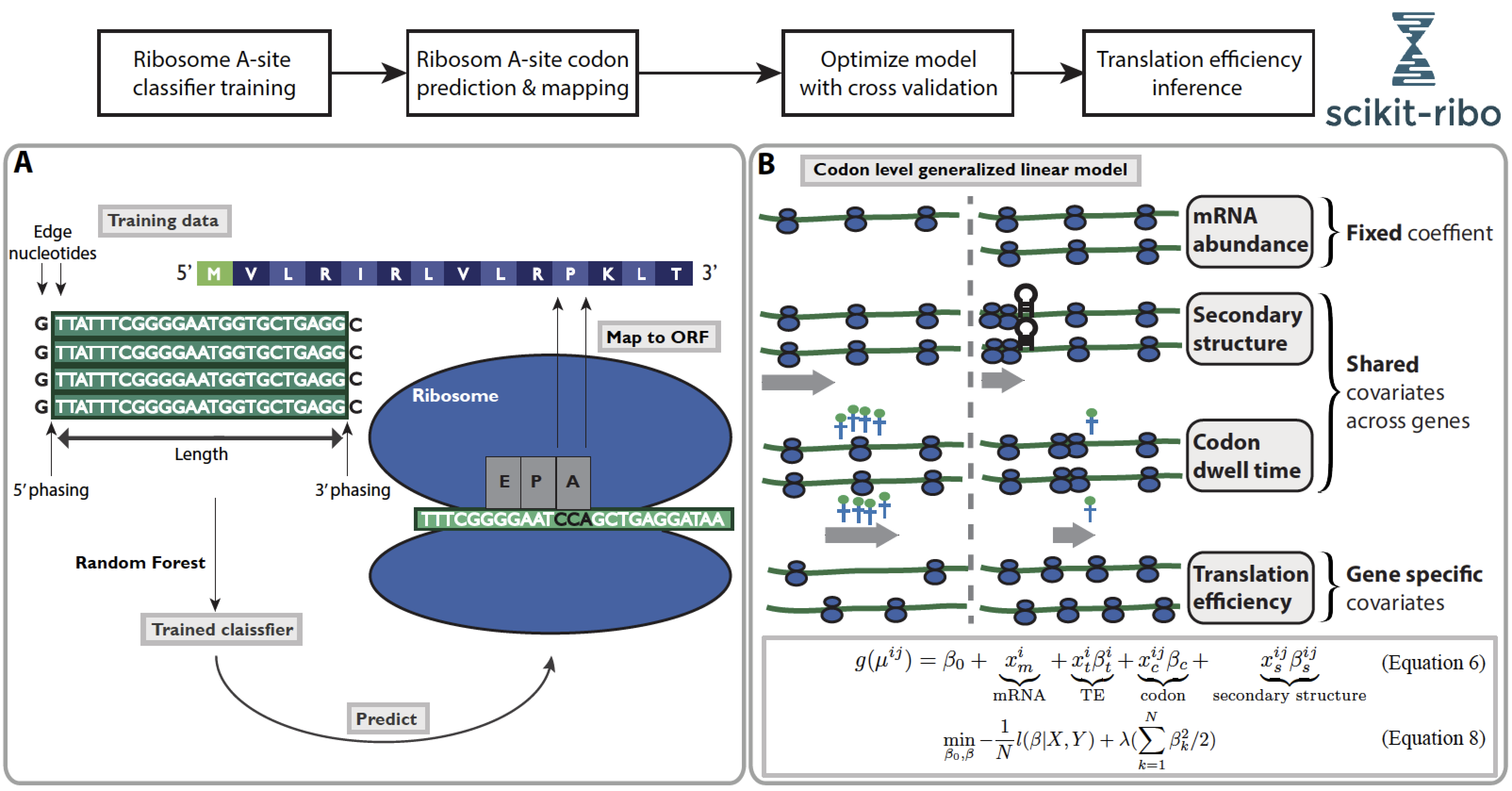
Scikit-ribo has two major modules:
- Ribosome A-site location prediction using random forest with recursive feature selection
- Translation efficiency inference using a codon-lvel generalized linear model with ridge penalty
A complete analysis with scikit-ribo has two major procedures:
- The data pre-processing step to prepare the ORFs, codons for a genome:
scikit-ribo-build.py - The actual model training and fitting:
scikit-ribo-run.py
Detailed workflow¶
Inputs¶
- The alignment of Riboseq reads (bam)
- Gene-level quantification of RNA-seq reads (from either Salmon or Kallisto)
- A gene annotation file (gtf)
- A reference genome for the model organism of interest (fasta)
Output¶
- Translation efficiency estimates for the genes
- Translation elongation rate for 61 sense codons
- Ribosome profile plots for each gene
- Diagnostic plots of the models
Cite¶
Fang et al, “Scikit-ribo: Accurate inference and robust modelling of translation dynamics at codon resolution” (Preprint coming up)
Contact¶
Han Fang
Stony Brook University & Cold Spring Harbor Laboratory
Email: hanfang.cshl@gmail.com